Life, for most of us, ends far too soon — hence the effort by biomedical researchers to find ways to delay the aging process and extend our stay on Earth. But there’s a paradox at the heart of the science of aging: The vast majority of research focuses on fruit flies, nematode worms and laboratory mice, because they’re easy to work with and lots of genetic tools are available. And yet, a major reason that geneticists chose these species in the first place is because they have short lifespans. In effect, we’ve been learning about longevity from organisms that are the least successful at the game.
Today, a small number of researchers are taking a different approach and studying unusually long-lived creatures — ones that, for whatever evolutionary reasons, have been imbued with lifespans far longer than other creatures they’re closely related to. The hope is that by exploring and understanding the genes and biochemical pathways that impart long life, researchers may ultimately uncover tricks that can extend our own lifespans, too.
Everyone has a rough idea of what aging is, just from experiencing it as it happens to themselves and others. Our skin sags, our hair goes gray, joints stiffen and creak — all signs that our components — that is, proteins and other biomolecules — aren’t what they used to be. As a result, we’re more prone to chronic diseases such as cancer, Alzheimer’s and diabetes — and the older we get, the more likely we are to die each year. “You live, and by living you produce negative consequences like molecular damage. This damage accumulates over time,” says Vadim Gladyshev, who researches aging at Harvard Medical School. “In essence, this is aging.”
This happens faster for some species than others, though — the clearest pattern is that bigger animals tend to live longer lives than smaller ones. But even after accounting for size, huge differences in longevity remain. A house mouse lives just two or three years, while the naked mole rat, a similar-sized rodent, lives more than 35. Bowhead whales are enormous — the second-largest living mammal — but their 200-year lifespan is at least double what you’d expect given their size. Humans, too, are outliers: We live twice as long as our closest relatives, the chimpanzees.
Bats above average
Perhaps the most remarkable animal Methuselahs are among bats. One individual of Myotis brandtii, a small bat about a third the size of a mouse, was recaptured, still hale and hearty, 41 years after it was initially banded. That is especially amazing for an animal living in the wild, says Emma Teeling, a bat evolutionary biologist at University College Dublin who coauthored a review exploring the value of bats in studying aging in the 2018 Annual Review of Animal Biosciences. “It’s equivalent to about 240 to 280 human years, with little to no sign of aging,” she says. “So bats are extraordinary. The question is, Why?”
There are actually two ways to think about Teeling’s question. First: What are the evolutionary reasons that some species have become long-lived, while others haven’t? And, second: What are the genetic and metabolic tricks that allow them to do that?
Answers to the first question, at least in broad brushstrokes, are becoming fairly clear. The amount of energy that a species should put toward preventing or repairing the damage of living depends on how likely an individual is to survive long enough to benefit from all that cellular maintenance. “You want to invest enough that the body doesn’t fall apart too quickly, but you don’t want to over-invest,” says Tom Kirkwood, a biogerontologist at Newcastle University in the UK. “You want a body that has a good chance of remaining in sound condition for as long as you have a decent statistical probability to survive.”

This implies that a little scurrying rodent like a mouse has little to gain by investing much in maintenance, since it will probably end up as a predator’s lunch within a few months anyway. That low investment means it should age more quickly. In contrast, species such as whales and elephants are less vulnerable to predation or other random strokes of fate and are likely to survive long enough to reap the benefits of better-maintained cellular machinery. It’s also no surprise that groups such as birds and bats — which can escape enemies by flying — tend to live longer than you’d expect given their size, Kirkwood says. The same would apply for naked mole rats, which live their lives in subterranean burrows where they are largely safe from predators.
But the question that researchers most urgently want to answer is the second one: How do long-lived species manage to delay aging? Here, too, the outline of an answer is beginning to emerge as researchers compare species that differ in longevity. Long-lived species, they’ve found, accumulate molecular damage more slowly than shorter-lived ones do. Naked mole rats, for example, have an unusually accurate ribosome, the cellular structure responsible for assembling proteins. It makes only a tenth as many errors as normal ribosomes, according to a study led by Vera Gorbunova, a biologist at the University of Rochester. And it’s not just mole rats: In a follow-up study comparing 17 rodent species of varying longevity, Gorbunova’s team found that the longer-lived species, in general, tended to have more accurate ribosomes.
The proteins of naked mole rats are also more stable than those of other mammals, according to research led by Rochelle Buffenstein, a comparative gerontologist at Calico, a Google spinoff focused on aging research. Cells of this species have greater numbers of a class of molecules called chaperones that help proteins fold properly. They also have more vigorous proteasomes, structures that dispose of defective proteins. Those proteasomes become even more active when faced with oxidative stress, reactive chemicals that can damage proteins and other biomolecules; in contrast, the proteasomes of mice become less efficient, thus allowing damaged proteins to accumulate and impair the cell’s workings.

DNA, too, seems to be maintained better in longer-lived mammals. When Gorbunova’s team compared the efficiency with which 18 rodent species repaired a particular kind of damage (called a double-strand break) in their DNA molecules, they found that species with longer lifespans, such as naked mole rats and beavers, outperformed shorter-lived species such as mice and hamsters. The difference was largely due to a more powerful version of a gene known as Sirt6, which was already known to affect lifespan in mice.
Watching the “epigenetic clock”
But it’s not just the genes themselves that suffer as animals age — so does their pattern of activation. An important way that cells turn genes on and off at the right time and place is by attaching chemical tags called methyl groups to sites that control gene activity. But these tags — also known as epigenetic marks — tend to get more random over time, leading gene activity to become less precise. In fact, geneticist Steve Horvath of UCLA and his colleagues have found that by assessing the status of a set of almost 800 methylation sites scattered around the genome, they can reliably estimate an individual’s age relative to the maximum lifespan of its species. This “epigenetic clock” holds for all the 192 species of mammals that Horvath’s team has looked at so far.
Notably, the epigenetic marks of longer-lived mammals take longer to degrade, which presumably means that their genes maintain youthful activity longer. In bats, for example, the longest-lived bats often have the slowest rate of change in methylations, while shorter-lived species change more quickly (see diagram).
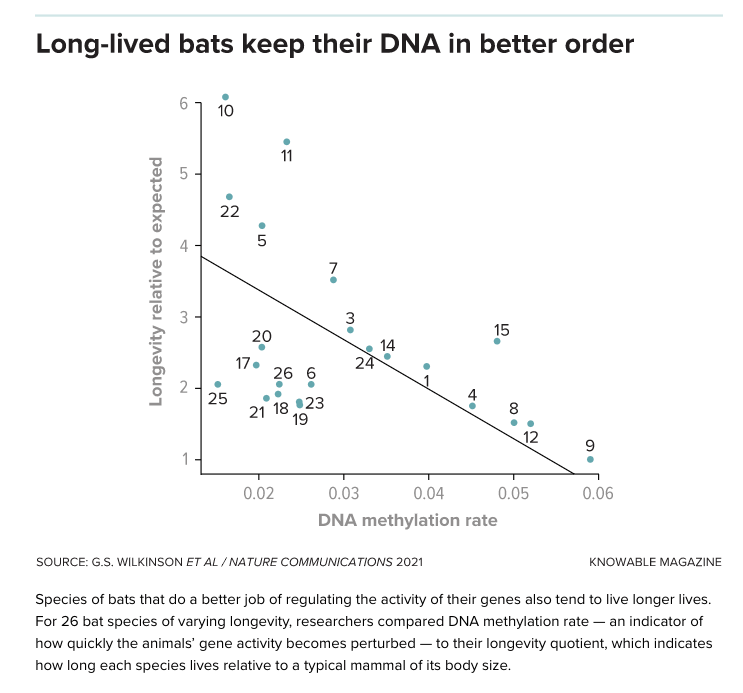
As he digs deeper, Horvath is finding that certain methylation sites may predict a species’ lifespan regardless of the age at which he samples them. “To me, this is a miracle,” he says. “Let’s say you go into the jungle and find a new species — could be a new bat or any other mammal. I can tell you pretty accurately the maximum lifespan of the species.” The methylation clues also predict maximum lifespan for dog breeds, which may emerge as an important study organism for aging (see sidebar: “What Rover knows”). These lifespan-related methylations tend to be associated with genes related to development, Horvath finds, though more detailed connections have yet to be worked out. He hopes that this work, which is not yet published, can eventually point the researchers toward genes that are key for regulating lifespan and aging.
Improvements in molecular techniques are already giving researchers more powerful tools to tease out the ways in which extraordinarily long-lived organisms may differ from the ordinary. One promising technique involves sequencing not the DNA in cells, but the messenger RNA. Individual genes are copied into mRNA as the first step in producing proteins, so mRNA sequencing reveals which genes in the genome are active at any given moment. This profile — referred to as the transcriptome — gives a more dynamic view of a cell’s activity than just listing the genes in the genome.
Gladyshev’s team, for example, sequenced the transcriptomes of cells from the liver, kidney and brain of 33 species of mammals, then looked for patterns that correlated with lifespan. They found plenty, including differences in activity levels of many genes involved in cellular maintenance functions such as DNA repair, antioxidant defense and detoxification.
Other paths to old age
More recently, Teeling’s team studied Myotis myotis bats from five roosts in France for eight years, capturing each bat every year and taking small samples of blood for transcriptome sequencing. This allowed them to track how the bats’ transcriptomes changed as they aged and compare the process to that of mice, wolves and people — the only other species for which similar long-term transcriptome data were available. “As the bats age,” Teeling wondered, “do they show the same dysregulation that we would show as we age?”
The answer, it turned out, was no. Whereas the other mammals produced fewer and fewer mRNA molecules related to maintenance functions such as DNA repair and protein stability the older they got, the bats did not. Instead, their maintenance systems seemed to get stronger as they got older, producing more repair-related mRNAs.
Skeptics note that conclusive evidence is still lacking, because the presence of more mRNA molecules does not necessarily mean more effective maintenance. “It’s an important first step, but it’s only that,” says Steven Austad, a biogerontologist at the University of Alabama, Birmingham. Still, the fact that the analysis identified processes that were already linked to longevity, such as DNA repair and protein maintenance, suggests that other genes flagged by this method could be solid leads: “We could then go look at new pathways that we haven’t yet explored,” Teeling says. In particular, the team found 23 genes that become much more active with age in bats but less active in other mammals. They are now looking at these genes with great interest, in the hopes of discovering new levers to alter the course of aging.
One of the principles beginning to emerge from comparative studies of aging is that different species may follow different paths to longevity. All long-lived mammals need to delay the onset of cancer, for example. Elephants do this by having multiple copies of key tumor-suppressing genes, so that every cell has backups if one gene breaks during the wear and tear of life. Naked mole rats, on the other hand, gain cancer resistance from an unusual molecule involved in sticking cells together, while bowhead whales have amped up their DNA repair pathways.

Geroscientists tend to view this diversity of solutions as an aid in their quest, not a problem. “That makes our job more difficult, but actually more interesting,” says Austad. “By studying the diversity of ways to achieve slow aging and long life, I think we’re more likely to stumble on things that are more easily translated to humans.”
Can we live longer, healthier lives by learning how to be more like naked mole rats, bats and bowhead whales? Not anytime soon — but the early results from research on these animal Methuselahs show definite promise.
View the original article here











